Reading the BEDMAP CSV files#
Author: Alice Fremand (@almand)
Date: 30/09/2022
Aim#
The goal of this tutorial is to show how the BEDMAP CSV files have been checked geospatially. The code can also be run to create shapefiles or geopackages or directly check the data geospatially.
The data#
The BEDMAP CSV files are available for downmoad from the UK Polar data Centre:
The code#
Virtual environment#
For the code to run, it is important to install the correct dependancies and libraries. In particular the following libraries are crucial for the code to be run:
geopandas
numpy
Scipy
math
Note: It is recommended to install geopandas first as it will upload most of the needed libraries at the same time without interoperability issues.
To set up the virtual environment with Conda:#
>conda create -n geo_env
>conda activate geo_env
>conda config --env --add channels conda-forge
>conda config --env --set channel_priority strict
>conda install python=3 geopandas
To set up the virtual environment on the SAN:#
>module load python/conda3
Then in the folder where you have your code, you need to launch:
>python3 -m venv geo_env
It will create a folder with all the environment for python. To activate the virtual environment you need to lauch it:
>source venv/bin/activate.csh
>source activate geo_env
You need to make sure that [geo_env] appears before your name on the machine. That means that you are using the virtual environment Then you need to upgrade pip which is the command that install the packages
>python3 -m pip install --upgrade pip
And install the other libraries
>python3 -m pip install geopandas
In this tutorial, the virtual environment is already set up. The list of the current libraries loaded is given in the list below.
pip list
Package Version
----------------------------- --------------
accessible-pygments 0.0.5
affine 2.4.0
alabaster 0.7.16
anyio 4.8.0
argon2-cffi 23.1.0
argon2-cffi-bindings 21.2.0
arrow 1.3.0
asttokens 3.0.0
async-lru 2.0.4
attrs 25.1.0
babel 2.17.0
beautifulsoup4 4.13.3
bleach 6.2.0
branca 0.8.1
Brotli 1.1.0
cached-property 1.5.2
Cartopy 0.24.0
certifi 2025.1.31
cffi 1.17.1
cftime 1.6.4
charset-normalizer 3.4.1
click 8.1.8
click-plugins 1.1.1
cligj 0.7.2
colorama 0.4.6
comm 0.2.2
contourpy 1.3.1
cycler 0.12.1
debugpy 1.8.12
decorator 5.2.1
defusedxml 0.7.1
docutils 0.21.2
exceptiongroup 1.2.2
executing 2.1.0
fastjsonschema 2.21.1
folium 0.19.4
fonttools 4.56.0
fqdn 1.5.1
geopandas 1.0.1
greenlet 3.1.1
h11 0.14.0
h2 4.2.0
hpack 4.1.0
httpcore 1.0.7
httpx 0.28.1
hyperframe 6.1.0
idna 3.10
imagesize 1.4.1
importlib_metadata 8.6.1
importlib_resources 6.5.2
ipykernel 6.29.5
ipython 8.32.0
isoduration 20.11.0
jedi 0.19.2
Jinja2 3.1.5
joblib 1.4.2
json5 0.10.0
jsonpointer 3.0.0
jsonschema 4.23.0
jsonschema-specifications 2024.10.1
jupyter-book 1.0.4.post0
jupyter-cache 1.0.1
jupyter_client 8.6.3
jupyter_core 5.7.2
jupyter-events 0.12.0
jupyter-lsp 2.2.5
jupyter_server 2.15.0
jupyter_server_terminals 0.5.3
jupyterlab 4.3.5
jupyterlab_pygments 0.3.0
jupyterlab_server 2.27.3
kiwisolver 1.4.7
latexcodec 2.0.1
linkify-it-py 2.0.3
mapclassify 2.8.1
markdown-it-py 3.0.0
MarkupSafe 3.0.2
matplotlib 3.10.0
matplotlib-inline 0.1.7
mdit-py-plugins 0.4.2
mdurl 0.1.2
mistune 3.1.2
munkres 1.1.4
myst-nb 1.2.0
myst-parser 3.0.1
nbclient 0.10.2
nbconvert 7.16.6
nbformat 5.10.4
nest_asyncio 1.6.0
netCDF4 1.7.2
networkx 3.4.2
notebook_shim 0.2.4
numpy 2.2.3
overrides 7.7.0
packaging 24.2
pandas 2.2.3
pandocfilters 1.5.0
parso 0.8.4
pickleshare 0.7.5
pillow 11.1.0
pip 25.0.1
pkgutil_resolve_name 1.3.10
platformdirs 4.3.6
prometheus_client 0.21.1
prompt_toolkit 3.0.50
psutil 7.0.0
pure_eval 0.2.3
pybtex 0.24.0
pybtex-docutils 1.0.3
pycparser 2.22
pydata-sphinx-theme 0.15.4
Pygments 2.19.1
pyogrio 0.10.0
pyparsing 3.2.1
pyproj 3.7.1
pyshp 2.3.1
PySocks 1.7.1
python-dateutil 2.9.0.post0
python-json-logger 2.0.7
pytz 2024.1
pywin32 307
pywinpty 2.0.15
PyYAML 6.0.2
pyzmq 26.2.1
rasterio 1.4.3
referencing 0.36.2
requests 2.32.3
rfc3339_validator 0.1.4
rfc3986-validator 0.1.1
rpds-py 0.23.1
scikit-learn 1.6.1
scipy 1.15.2
Send2Trash 1.8.3
setuptools 75.8.2
shapely 2.0.7
six 1.17.0
sniffio 1.3.1
snowballstemmer 2.2.0
snuggs 1.4.7
soupsieve 2.5
Sphinx 7.4.7
sphinx-book-theme 1.1.4
sphinx-comments 0.0.3
sphinx-copybutton 0.5.2
sphinx_design 0.6.1
sphinx_external_toc 1.0.1
sphinx-jupyterbook-latex 1.0.0
sphinx-multitoc-numbering 0.1.3
sphinx-thebe 0.3.1
sphinx-togglebutton 0.3.2
sphinxcontrib-applehelp 2.0.0
sphinxcontrib-bibtex 2.6.3
sphinxcontrib-devhelp 2.0.0
sphinxcontrib-htmlhelp 2.1.0
sphinxcontrib-jsmath 1.0.1
sphinxcontrib-qthelp 2.0.0
sphinxcontrib-serializinghtml 1.1.10
SQLAlchemy 2.0.38
stack_data 0.6.3
tabulate 0.9.0
terminado 0.18.1
threadpoolctl 3.5.0
tinycss2 1.4.0
tomli 2.2.1
tornado 6.4.2
traitlets 5.14.3
types-python-dateutil 2.9.0.20241206
typing_extensions 4.12.2
typing_utils 0.1.0
tzdata 2025.1
uc-micro-py 1.0.3
uri-template 1.3.0
urllib3 2.2.2
wcwidth 0.2.13
webcolors 24.11.1
webencodings 0.5.1
websocket-client 1.8.0
wget 3.2
win_inet_pton 1.1.0
xarray 2025.1.2
xyzservices 2025.1.0
zipp 3.21.0
Note: you may need to restart the kernel to use updated packages.
Upload the modules#
geopandas: used to create geodataframe and easily save the result to shapefiles or geopackages.
pandas: used to read the csv file
Other modules: mathplotlib
import pandas as pd
import geopandas as gpd
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
Import the csv file#
Before starting, the BEDMAP CSV files need to be downloaded from the UK Polar Data Centre Repository. In this example, we use the ‘AWI_2014_GEA-IV_AIR_BM3’ file located in the following folder: 'D:/BEDMAP/AWI_2015_GEA-DML_AIR_BM3.csv'
CSV_file = 'AWI_2015_GEA-DML_AIR_BM3.csv'
The pandas module is used to open the file. The command pd.read_csv(input_file) can be used to read the BEDMAP CSV file. To skip the metadata at the top of the file, we use the skiprows argument as follows:
data = pd.read_csv(CSV_file, skiprows=18, low_memory=False)
By using the read_csv() command, the data is directly saved in a dataframe. We can have a look at the top lines by running the following:
data.head()
| trajectory_id | trace_number | longitude (degree_east) | latitude (degree_north) | date | time_UTC | surface_altitude (m) | land_ice_thickness (m) | bedrock_altitude (m) | two_way_travel_time (m) | aircraft_altitude (m) | along_track_distance (m) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 20162002_01 | -9999 | -5.7951 | -71.0262 | 2015-12-15 | 12:43:36 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 0 |
| 1 | 20162002_01 | -9999 | -5.7937 | -71.0264 | 2015-12-15 | 12:43:37 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 56 |
| 2 | 20162002_01 | -9999 | -5.7871 | -71.0275 | 2015-12-15 | 12:43:42 | -9999 | 456.3 | -9999 | 0.000005 | -9999 | 325 |
| 3 | 20162002_01 | -9999 | -5.7824 | -71.0282 | 2015-12-15 | 12:43:45 | -9999 | 452.6 | -9999 | 0.000005 | -9999 | 512 |
| 4 | 20162002_01 | -9999 | -5.7729 | -71.0296 | 2015-12-15 | 12:43:51 | -9999 | 445.3 | -9999 | 0.000005 | -9999 | 891 |
data.dtypes
trajectory_id object
trace_number int64
longitude (degree_east) float64
latitude (degree_north) float64
date object
time_UTC object
surface_altitude (m) int64
land_ice_thickness (m) float64
bedrock_altitude (m) int64
two_way_travel_time (m) float64
aircraft_altitude (m) int64
along_track_distance (m) int64
dtype: object
data.tail()
| trajectory_id | trace_number | longitude (degree_east) | latitude (degree_north) | date | time_UTC | surface_altitude (m) | land_ice_thickness (m) | bedrock_altitude (m) | two_way_travel_time (m) | aircraft_altitude (m) | along_track_distance (m) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 23377 | 20162020_04 | -9999 | -7.3021 | -70.6626 | 2016-01-03 | 14:51:27 | -9999 | 251.8 | -9999 | 0.000003 | -9999 | 161042 |
| 23378 | 20162020_04 | -9999 | -7.3057 | -70.6625 | 2016-01-03 | 14:51:28 | -9999 | 252.4 | -9999 | 0.000003 | -9999 | 161176 |
| 23379 | 20162020_04 | -9999 | -7.3119 | -70.6624 | 2016-01-03 | 14:51:32 | -9999 | 251.6 | -9999 | 0.000003 | -9999 | 161406 |
| 23380 | 20162020_04 | -9999 | -7.3181 | -70.6623 | 2016-01-03 | 14:51:35 | -9999 | 252.5 | -9999 | 0.000003 | -9999 | 161636 |
| 23381 | 20162020_04 | -9999 | -7.3234 | -70.6622 | 2016-01-03 | 14:51:38 | -9999 | 253.0 | -9999 | 0.000003 | -9999 | 161832 |
Convert the file geospatially#
The geopandas module is then used to convert the data to a geodataframe. It will convert the latitude/longitude to points.
To do that, you will need to identify the specific header used for longitude and latitude in the CSV file.
Here, it is Longitude_decimal_degrees and Latitude_decimal_degrees
gdf = gpd.GeoDataFrame(data, geometry=gpd.points_from_xy(data['longitude (degree_east)'], data['latitude (degree_north)']))
We can check that the conversion has been done:
gdf.head()
| trajectory_id | trace_number | longitude (degree_east) | latitude (degree_north) | date | time_UTC | surface_altitude (m) | land_ice_thickness (m) | bedrock_altitude (m) | two_way_travel_time (m) | aircraft_altitude (m) | along_track_distance (m) | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 20162002_01 | -9999 | -5.7951 | -71.0262 | 2015-12-15 | 12:43:36 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 0 | POINT (-5.7951 -71.0262) |
| 1 | 20162002_01 | -9999 | -5.7937 | -71.0264 | 2015-12-15 | 12:43:37 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 56 | POINT (-5.7937 -71.0264) |
| 2 | 20162002_01 | -9999 | -5.7871 | -71.0275 | 2015-12-15 | 12:43:42 | -9999 | 456.3 | -9999 | 0.000005 | -9999 | 325 | POINT (-5.7871 -71.0275) |
| 3 | 20162002_01 | -9999 | -5.7824 | -71.0282 | 2015-12-15 | 12:43:45 | -9999 | 452.6 | -9999 | 0.000005 | -9999 | 512 | POINT (-5.7824 -71.0282) |
| 4 | 20162002_01 | -9999 | -5.7729 | -71.0296 | 2015-12-15 | 12:43:51 | -9999 | 445.3 | -9999 | 0.000005 | -9999 | 891 | POINT (-5.7729 -71.0296) |
Determine the variables to show in the shapefile/geopackage#
All the variables that are present in the dataframe will be shown in the final shapefile or geopackage. We might want to remove the columns that we don’t want. For instance, here all the line_ID values are equal to -9999, so this information is missing. To delete this column, we just need to do the following:
gdf = gdf.drop(columns=['trajectory_id'])
gdf.head()
| trace_number | longitude (degree_east) | latitude (degree_north) | date | time_UTC | surface_altitude (m) | land_ice_thickness (m) | bedrock_altitude (m) | two_way_travel_time (m) | aircraft_altitude (m) | along_track_distance (m) | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -9999 | -5.7951 | -71.0262 | 2015-12-15 | 12:43:36 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 0 | POINT (-5.7951 -71.0262) |
| 1 | -9999 | -5.7937 | -71.0264 | 2015-12-15 | 12:43:37 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 56 | POINT (-5.7937 -71.0264) |
| 2 | -9999 | -5.7871 | -71.0275 | 2015-12-15 | 12:43:42 | -9999 | 456.3 | -9999 | 0.000005 | -9999 | 325 | POINT (-5.7871 -71.0275) |
| 3 | -9999 | -5.7824 | -71.0282 | 2015-12-15 | 12:43:45 | -9999 | 452.6 | -9999 | 0.000005 | -9999 | 512 | POINT (-5.7824 -71.0282) |
| 4 | -9999 | -5.7729 | -71.0296 | 2015-12-15 | 12:43:51 | -9999 | 445.3 | -9999 | 0.000005 | -9999 | 891 | POINT (-5.7729 -71.0296) |
As you can see, the column Line_ID has been removed. We can also remove several variables at the same time:
gdf = gdf.drop(columns=[ 'trace_number', 'date', 'time_UTC'])
gdf.head()
| longitude (degree_east) | latitude (degree_north) | surface_altitude (m) | land_ice_thickness (m) | bedrock_altitude (m) | two_way_travel_time (m) | aircraft_altitude (m) | along_track_distance (m) | geometry | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -5.7951 | -71.0262 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 0 | POINT (-5.7951 -71.0262) |
| 1 | -5.7937 | -71.0264 | -9999 | 463.7 | -9999 | 0.000006 | -9999 | 56 | POINT (-5.7937 -71.0264) |
| 2 | -5.7871 | -71.0275 | -9999 | 456.3 | -9999 | 0.000005 | -9999 | 325 | POINT (-5.7871 -71.0275) |
| 3 | -5.7824 | -71.0282 | -9999 | 452.6 | -9999 | 0.000005 | -9999 | 512 | POINT (-5.7824 -71.0282) |
| 4 | -5.7729 | -71.0296 | -9999 | 445.3 | -9999 | 0.000005 | -9999 | 891 | POINT (-5.7729 -71.0296) |
Setting up the coordinate system#
It is important to then set the coordinate system. Here the WGS84 coordinate system is used, it corresponds to the EPSG: 4326.
gdf = gdf.set_crs("EPSG:4326")
With geopandas, it is also possible to convert the data to another coordinate system and project it. You just need to know the EPSG ID of the output coordinate system. Here is how to convert the data to the stereographic geographic system.
gdf = gdf.to_crs("EPSG:3031")
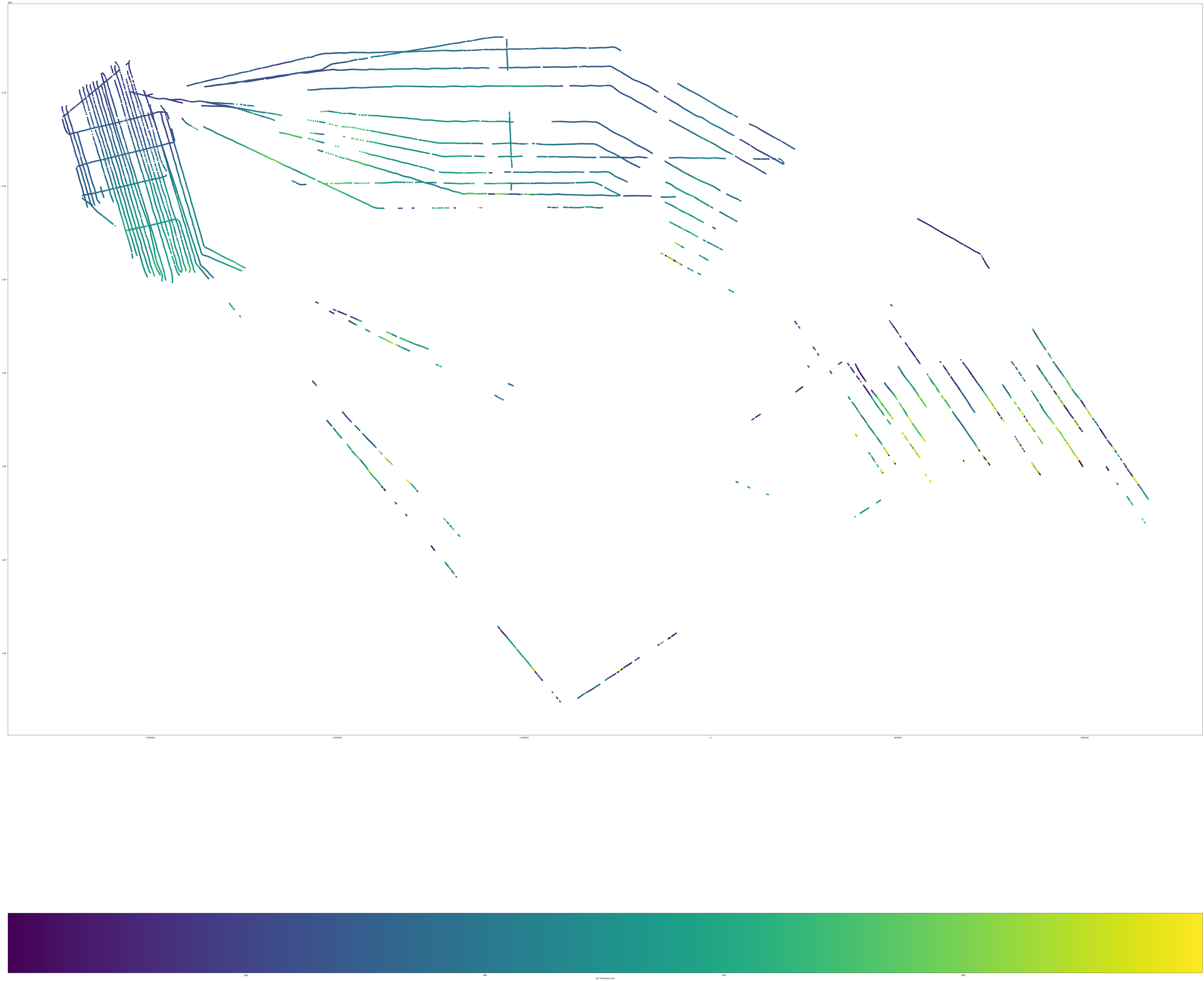
Plotting the data#
plt.rcParams["figure.figsize"] = (100,100)
fig, ax = plt.subplots(1, 1)
fig=plt.figure(figsize=(100,100), dpi= 100, facecolor='w', edgecolor='k')
gdf.plot(column='land_ice_thickness (m)', ax=ax, legend=True, legend_kwds={'label': "Ice Thickness (m)",'orientation': "horizontal"})
<Axes: >
<Figure size 10000x10000 with 0 Axes>
Saving the data to geopackages or shapefile#
To save the data to geopackage or shapefile, we only need to use the .to_file() command from geopandas module. Then we specify the driver, that will specify the type of output - geopackage or shapefile.
gdf.to_file('point.gpkg', layer='Points', driver="GPKG")
# or for shapefile:
#gdf.to_file('point.shp', driver="ESRI Shapefile")

